Schizophrenia is a complex psychiatric disorder with polygenic and potential environmental contributions to its etiology. In the last few decades, genetic risk factors have been identified but their biological mechanisms remain largely unknown. A new study published in the journal Cell Reports by a team from the International Research Center for Neurointelligence, The University of Tokyo Institutes for Advanced Study, found a novel connection between a gene called Setd1a and impairments in synaptic function and social behavior in schizophrenia. The results are another step in understanding the origins of mental illness and social intelligence.
Setd1a belongs to a class of nuclear genes encoding histone methyltransferases that regulate the epigenome, the essential packaging material for DNA that also serves as a scaffold for the many regulatory proteins and small RNAs that orchestrate gene expression in our cells, including neurons in the brain’s neocortex that control complex behaviors and diseases. Mutations that incapacitate the expression or function of SETD1A have been identified in some schizophrenia patients and their presence results in an increased disease risk, but how these mutations work was unclear. How does the Setd1a epigenome regulate neuronal function?
The team generated Setd1a mutant mice containing a unique mutation found in a human patient and subjected animals to a behavioral screen. Interestingly, the mice showed defects in behavior including sociality, working memory, and novel object recognition. Synaptic measurements revealed a deficit of synaptic transmission in a particular cell type, the layer 2/3 excitatory pyramidal cell in the medial prefrontal cortex (mPFC). This observation was accompanied by a reduced release probability of glutamate and fewer synaptic spines. To further examine the role of mPFC in excitatory synaptic transmission and behavior the team generated mice with a reduction of Setd1a in mPFC and confirmed the decrease in synaptic function and sociality.
The observed defects in synaptic structure and function resulting in impaired social behavior suggested that Setd1a might regulate the epigenome leading to changes in the expression of a cluster of synaptic and disease related genes. They sampled tissue from the mPFC of mutant mice and conducted RNA sequence profiling, a method that can identify most of the genes that are regulated under a particular condition, in this case, those due to the loss of Setd1a. The results revealed increases and decreases in the levels of hundreds of genes including those related to synaptic functions, and developmental brain diseases and disorders. Further study of this Setd1a genetic network may help to understand the schizophrenic patient brain.
Correspondent: Charles Yokoyama, Ph.D., IRCN Science Writing Core
Reference: Nagahama, K., Sakoori, K., Watanabe, T., Kishi, Y., Kawaji, K., Koebis, M., Nakao, K., Gotoh, Y., Aiba, A., Uesaka, N., Kano, M. (2020) Setd1a Insufficiency in Mice Attenuates Excitatory Synaptic Function and Recapitulates Schizophrenia-related Behavioral Abnormalities. Cell Reports, Online publication: September 14, 2020, DOI: 10.1016/j.celrep.2020.108126
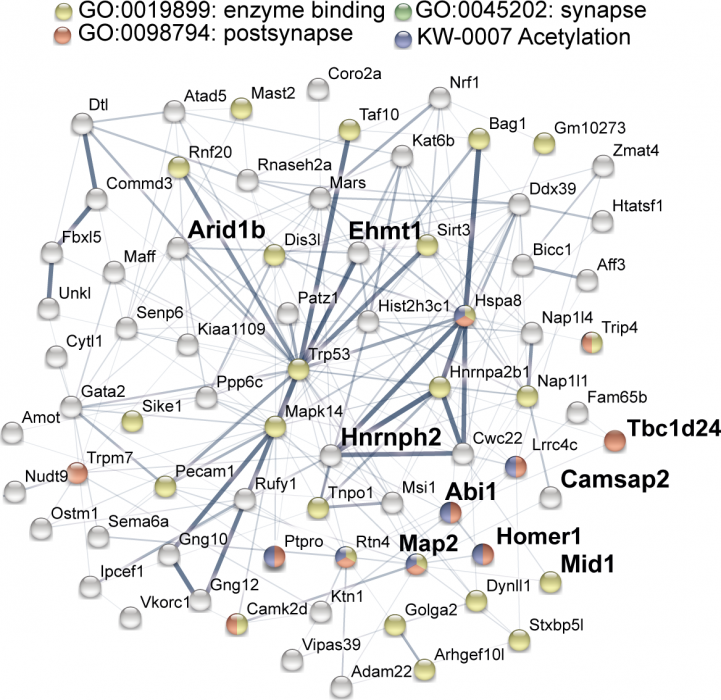
Figure: Down-regulated protein network in Setd1a +/- mice indicating links between synaptic functions and epigenomic modifications. Bold represents neurodevelopmental disorder genes.
Media Contact: The author is available for interviews in English.
Professor Masanobu Kano, M.D., Ph.D.
International Research Center for Neurointelligence (IRCN)
The University of Tokyo Institutes for Advanced Study
Mayuki Satake
Public Relations
International Research Center for Neurointelligence (IRCN)
The University of Tokyo Institutes for Advanced Study
pr@ircn.jp


