The clinical diagnosis of a disease is based on a doctor’s assessment after seeing a patient’s overt symptoms and running confirmatory laboratory tests. However, the development of a disease obviously begins far earlier and novel methods to predict the so-called pre-disease state based on physiological measurements are of urgent medical and scientific interest.
In a recent study from a team of researchers led by Professor Kazuyuki Aihara of the International Research Center for Neurointelligence, University of Tokyo Institutes for Advanced Study and Dr. Keiichi Koizumi of the University of Toyama, new insights into the measurement and mathematical theory of the pre-disease state for a common chronic disorder called metabolic syndrome modeled in mice were published in the journal Scientific Reports.
In a metabolic syndrome model, TSOD mice, animals developed overt symptoms at 8-10 weeks old that mirrored the human disorder, which is related to excess abdominal fat and can lead to various health problems including heart disease. In a mathematical theory developed previously by the Aihara laboratory called dynamical network biomarkers (DNB) the researchers expected that a pre-disease state should precede a transition to overt disease.
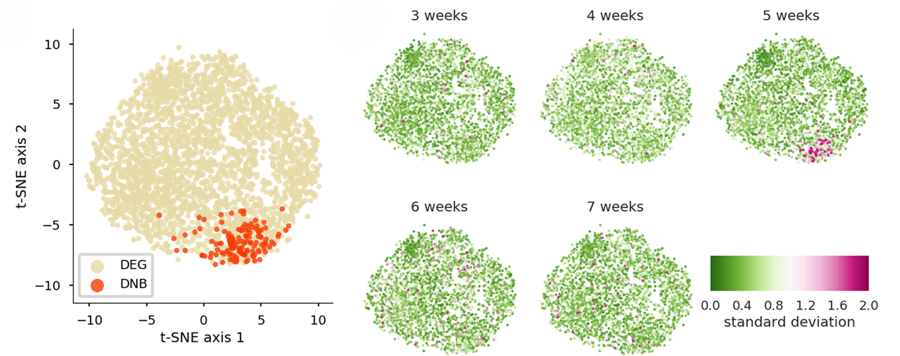
The team measured large scale gene expression using DNA microarrays to capture the genetic signature of possible pre-disease state at 5 weeks of age before the emergence of symptoms. They found that 147 genes comprised a set that could describe a 5 week old animal’s pre-disease state within the DNB theory. Although the results are promising, future studies will need to replicate them and deal with possible confounding conditions such as parallel changes in the microbiome of the adipose tissue where the samples were collected.
The findings add metabolic syndrome to a growing list of major diseases and conditions such as Alzheimer’s, diabetes, influenza, and cancer where the DNB theory has been shown to be useful in disease prediction. Future work will focus on psychiatric and other brain disorders. The hope is that a mathematical understanding of how human disease can be characterized by critical states and their transitions will allow earlier interventions in pervasive illnesses. More broadly, the DNB theory may be applied to any dynamic process to predict “tipping points”.
Correspondent: Charles Yokoyama, Ph.D., IRCN Science Writing Core
Reference: Koizumi, K., Oku, M., Hayashi, S., Inujima, A., Shibahara, N., Chen, L., Igarashi, Y., Tobe, K., Saito, S., Kadowaki, M., Aihara, K. (2019) Identifying pre-disease signals before metabolic syndrome in mice by dynamical network biomarkers. Scientific Reports 9:8767, DOI: 10.1038/s41598-019-45119-w
Figure
Spatiotemproral fluctuation patterns of the union set of 147 DNB genes within the larger differentially expressed population of 2665 genes. A T-distributed stochastic neighbor embedding (t-SNE) dimensionality reduction method demonstrated that most DNB genes were concentrated in one region at 5 weeks of age. The findings indicate that the DNB gene cluster may represent a unique biomarker for disease onset.
Media Contact: The author is available for interviews in English.
Professor Kazuyuki Aihara, Ph.D.
Institute of Industrial Science (IIS)
International Research Center for Neurointelligence (IRCN)
The University of Tokyo Institutes for Advanced Study
Mayuki Satake
Public Relations
International Research Center for Neurointelligence
The University of Tokyo
pr@ircn.jp


