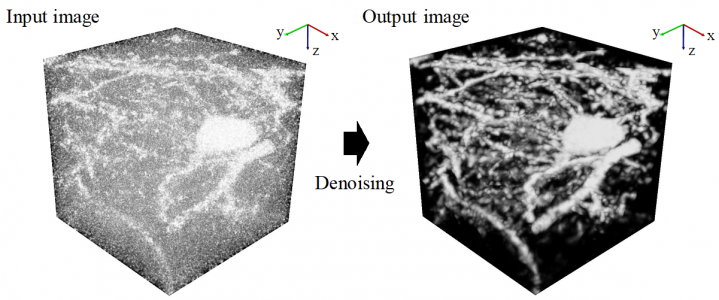
AI based on deep machine learning continues to serve as a platform for the development of new applications in biological research. With the development of more powerful microscopy systems coupled with novel molecular and cellular labels, the AI-driven deep analysis of biological images promises to unlock increasingly larger advances in understanding the structure and function of complex cellular populations such as those that comprise the brain’s neuronal networks. However, a current bottleneck in progress is the noise inherent in most images that prevents the acquisition of the type of clean, labelled images that deep networks require to better learn and predict new data. While some noise is a natural part of biology, the development of better methods to reduce the type of noise associated with the limitations of the microscopy methods themselves would help to reduce the unnecessary degradation of image quality and would be of immense help to researchers studying small effects in large datasets. Now, a cross-disciplinary research collaboration between a computation group in the Kyoto University Graduate School of Informatics and a biology lab from The University of Tokyo Graduate School of Medicine, both members of the International Research Center for Neurointelligence and led by Drs. Sehyung Lee and Shin Ishii, report the development of a new AI algorithm that represents a large improvement in the analysis of neuronal images, especially noisy 3-dimensional (3D) pictures from a subcellular microscopy method called 2-photon imaging. The study was recently published in the journal Neural Networks.
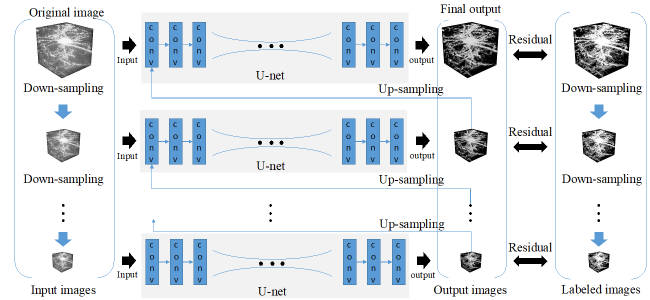
The Kyoto-based computational team set out to improve noise reduction and solve the unique problems of acquiring clearer images in 3D. Starting from a common deep learning model called a U-net convolutional neural network (CNN), they built a so-called stacked network architecture, with four U-nets arranged in tandem to process learning of images at coarse, medium, and fine scales, and termed Mu-net for “Multi-scale U-net”. The data were collected by the Tokyo-based biological team from whole mouse brains virally labelled in the posterior parietal cortex with fluorescent neuronal indicators, made transparent with a tissue clearing agent, and prepared for microscopy. To further enhance image quality, the researchers trained the Mu-net with another type of AI called a generative adversarial network (GAN) that enabled the images to become more robust to noise. Together, the Mu-net architecture with GAN training were successful in removing noise from images to a greater degree than other state of the art CNN-type models. Moreover, the Mu-net was constructed to enable end to end processing, a virtue in the machine learning field that avoids cumbersome pre- or post-image processing that might distort or bias the dataset. The authors suggest that Mu-net algorithms, involving a stacked architecture and adversarial training, might become standards in the field especially for the restoration of noisy 3D images and accurate mapping of 3D brain structure, resulting in improved deep learning analyses of single neurons or other biological elements. Future work will focus on applications of Mu-net to studies on brain and cognitive functions.
Correspondent: Charles Yokoyama, Ph.D., IRCN Science Writing Core
Reference: Lee, S., Negishi, M., Urakubo, H., Kasai, H., Ishii, S., (2020) Mu-net: Multi-scale U-net for two-photon microscopy image denoising and restoration. Neural Networks 125:92, DOI: 10.1016/j.neunet.2020.01.026
Media Contact: The author is available for interviews in English.
Sehyung Lee Ph.D.
Postdoctoral Researcher
Graduate School of Informatics
Kyoto University
Shin Ishii, Ph.D.
Affiliate Faculty
International Research Center for Neurointelligence (IRCN)
The University of Tokyo
Professor
Graduate School of Informatics
Kyoto University
Mayuki Satake
Public Relations
International Research Center for Neurointelligence (IRCN)
The University of Tokyo Institutes for Advanced Study
pr@ircn.jp
Figures: